A research team mainly led by Professor Jae Myoung Suh from the Graduate School of Medical Science and Engineering has developed a spatiotemporal method of identifying tissue-specific secretory proteins. The researchers have detected liver secretory proteins in the blood plasma of insulin-resistant mice using turboID, a bioengineered biotin ligase.
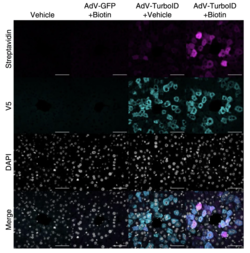
Secretory proteins are endocrine or exocrine proteins that are actively synthesized and transported out of a cell. As potential biomarkers, they are frequently profiled for disease diagnosis and serve as essential therapeutic targets. TurboID converts biotin (Vitamin H), a labeling reagent, to biotin-AMP, a reactive complex that readily binds to the secretory proteins. As compared to proteome data that could be derived from in vitro cell culture models, the new in vivo analyses of such proteins would give rise to a more reliable representation of internal conditions and physiological interactions. Researchers state that such improved methods would “resolve characteristics of tissue-specific secretory proteins along time and space dimensions”.
TurboID was introduced intravenously into mice by injecting an adenovirus into their tail veins, allowing the enzyme to anchor to the endoplasmic reticulum (ER) lumen of liver cells. Biotin was then administered to label the liver-specific proteins. The labeled proteins were separated from the supernatant of blood plasma through streptavidin affinity purification, and its molecular structure was analyzed through mass spectrometry. Serum albumin and pregnancy zone proteins were among the most abundant proteins discovered. Notably, the pattern of liver proteins derived in vivo had a low correlation with patterns previously derived from hepatocyte cell lines. Researchers agreed that the variation in results arised from the limited accuracy of in vitro analyses of proteins.
When researchers administered TurboID and biotin to insulin-resistant mice, the proteins commonly found in obese and diabetic human subjects were also detected from their blood plasma. The spectrometry illustrated elevated levels of Alpha-2-HS-glycoprotein (AHSG), Inter-Alpha-Trypsin Inhibitor Heavy Chain H1 (ITIH1), Afamin (AFM), and Beta-2-Glycoprotein (APOH), all of which are proven to impair normal glucose metabolism, thus causing diabetes.
The work of the research team is published in Nature Communications under the title, “Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice”. The study paves the way for exciting prospects in pathophysiology, including its uses in monitoring the progression of diseases and possible applications to other organ systems.

